Classification in Cryo-Electron Tomograms
SHREC 2020 Track

Motivation
There is a noticeable gap in knowledge about the organization of cellular life at the mesoscopic level. With the advent of the direct electron detectors and the associated resolution revolution, cryo-electron tomography (cryo-ET) has the potential to bridge this gap by simultaneously visualizing the cellular architecture and structural details of macromolecular assemblies, thee-dimensionally. The technique offers insights in key cellular processes and opens new possibilities for rational drug design. However, the biological samples are radiation sensitive, which limits the maximal resolution and signal-to-noise ratio. Innovation in computational methods remains key to derive biological information from the tomograms.
Task
In this SHREC track, we propose a task of localization and classification of biological particles in the cryo-electron tomogram volume. We provide physics-based simulation of cryo-electron tomograms and annotations for all of them except the test tomogram. We hope that this will enable researchers to try out different methods, including machine learning and statistical approaches. All 3D object retrieval and 3D electron microscopy experts interested in computational cryo-ET are welcome to participate.
Dataset
To provide participants with as accurate ground truth information as possible, we have created a physics-based simulator to generate cryo-electron tomograms.
The dataset consists of 10 tomograms, with 1nm/voxel resolution, with a size of 512x512x512 voxels. Each tomogram is packed with on average 2500 bio-particles of 12 uniformly distributed and rotated different proteins, various in size and structure, with the following PDB identificators:
For each tomogram but the test one, we also provide:
- Ground truth volume
- Ground truth tilt angle projections (using which the tomogram was constructed)
- Noise-free ground truth volume (ground truth without water and structural noise)
- Noise-free ground truth tilt angle projections
- Text file with locations and PDB ID of each particle
- Occupancy volumes (where each voxel contains particle ID of the particle (w.r.t. text file) or 0 if that’s not a particle)
- Class mask volumes (where each voxel contains class ID of the particle (w.r.t. text file) or 0 if that's not a particle)
Contest dataset has 9 annotated tomograms and 1 test tomogram.
Full dataset has 10 annotated tomograms.
Diff dataset contains only the difference between full and contest dataset. Download datasets Paper @ Computer&Graphics 3DOR journal Pre-print PDF
Registration
If you intend to participate in the track, please send us an email and mention your affiliation and co-authors.
This helps us keep track of the participants and plan accordingly. It also allows us to send you updates about the track.
Submission
From participants, no later than the deadline mentioned in the schedule, we expect results submitted along with a one-page description of the method used to generate them. Results should be presented as a .txt file containing the found particles, in the similar fashion to the ground truth text files. Data should be formatted in 4 columns: predicted class (PDB ID), estimated center X coordinates, estimated center Y coordinates, estimated center Z coordinates.
Evaluation
The main goal of the track is to localize and classify biological particles in the tomogram. The performance of methods will be evaluated solely on the test tomogram: the only tomogram for which ground truth is not provided. Following metrics will be measured and compared: precision, recall, F1 score. We intend to compare submitted results in two areas: localization (if a particle is found or not) and classification (if a found particle is correctly classified or not).
Changes from 2019
This year, we've improved our simulation process and are able to provide significantly more ground truth data to participants.
We replaced outdated structure with their successor (4b4t -> 4cr2).
This year SHREC track papers will follow a two-stage review process and, hopefully, will be published in the journal Computers & Graphics upon acceptance.
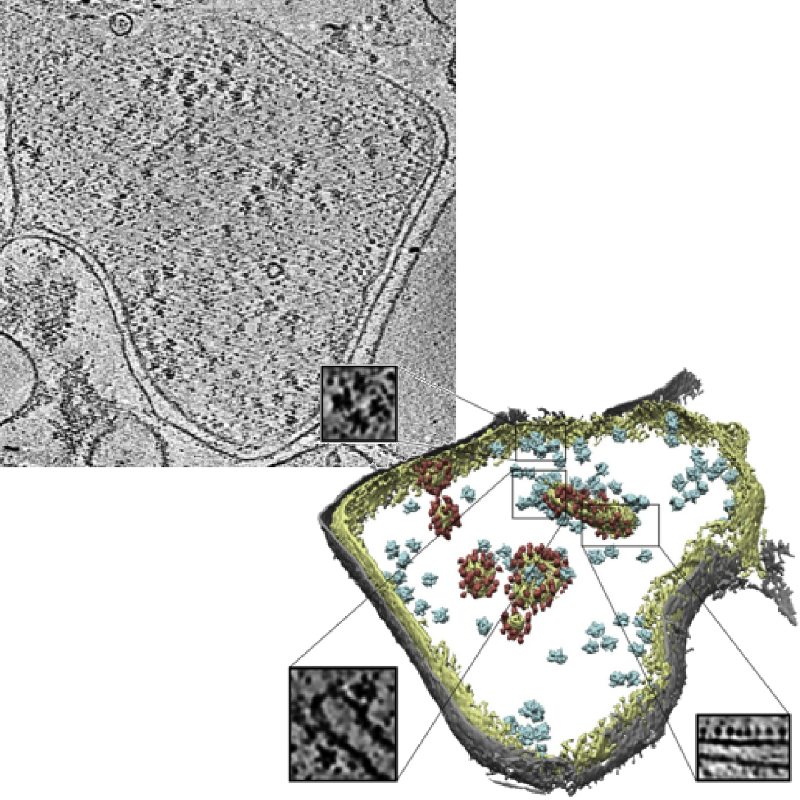
Slice of a tomogram and a 3D visualization obtained from the tomogram.
Image credits: Pfeffer S, Woellhaf MW, Herrmann JM, Förster F, Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nature communications 6:6019 (2015)
Organizers
- Ilja Gubins 1
- Marten Chaillet 2
- Gijs van der Schot 2
- Remco C. Veltkamp 1
- Friedrich G. Forster 2
2: Utrecht University, Department of Chemistry
Contact us
Schedule
The registration and submission deadlines are in AoE (Anywhere on Earth) timezone.
IMPORTANT: due to the worldwide spread of COVID-19, 3D Object Retrival Workshop organizers extended deadlines by 1 month, allowing us to extend track deadlines too.
| March 2 | Track announcement & dataset release |
April 2 |
Registration deadline |
April 20 |
Submission (results + one-page description) deadline |
| April 26 | Track organizers send track paper draft to participants for review |
May 15 |
Track organizers submit track paper for review |
May-July |
Track paper review process |
July 12 |
Camera-ready track paper is submitted for inclusion into the proceedings |
| September 4 | 3D Object Retrieval Workshop (w/ SHREC) |
Website last updated: Sep 5 2020 13:37 (CEST)